Most scientists regard the evidence for evolution as overwhelming. Thus, in their conviction that evolution has already been thoroughly and sufficiently documented, they sometimes fail to consider how new discoveries can be applied to support evolution. In this article, I draw together some discoveries of the past few years from my own field of molecular genetics. When these findings were initially reported, their implications for the creation-evolution controversy were not explicitly discussed; but they offer an interesting new twist to an old argument and provide evidence for evolution that is conceptually simple enough for the interested layperson to appreciate.
The new molecular evidence bears on a question which, in my opinion, represents one of the few cases in which a creationist argument had demonstrated logical consistency and had fought the evolutionary position to a deadlock. This is the question of how to interpret the similarities between modern species, especially the similarities observed at the molecular level. As we will see, the recent discoveries from molecular genetics resolve this deadlock in favor of evolution.
The Evolutionary View of Species Similarities
Consider first the interpretation of species similarities from the evolutionary viewpoint. Although present-day humans and gorillas may appear quite different from each other at first glance, their internal organs and physiological function are extremely similar: Just as the resemblance of two siblings suggests a common parentage, resemblance between species suggests common ancestors. Evolutionists believe that humans, gorillas, and chimpanzees evolved from a common ancestor—an apelike creature that lived perhaps five to ten million years ago, rather recently on the geological time scale. (The thought that humans and apes might share a common ancestor seems particularly unacceptable to creationists because of theological implications and the clear contradiction to the biblical account of human creation.) Species less similar to humans than are apes—mice, for example—are believed to have branched off millions of years earlier from a common primitive mammalian ancestor. Evolutionary family tree diagrams that express such relationships between species have been constructed by evolutionary biologists by analyzing similarities of present-day organisms. In many cases, fossilized remains of extinct species can be used to support the features of such evolutionary trees; fossil evidence will not, however, be discussed in this article.
Another extensive source of data that has been of major importance in constructing tree diagrams is the species comparison of proteins. Proteins are large biological molecules made of subunits called amino acids that are attached to one another in chains, like the cars of a train. There are twenty different kinds of amino acids used in proteins, and most proteins contain hundreds of these subunits. Each protein has a specific number and sequence of amino acids, and this sequence determines what properties that protein will have. The sequence information specifying the structure of each protein is stored in "blueprint" form in the organism's genes. Biochemists can purify proteins and learn the exact sequence of their amino acids. Considerable effort has gone into comparing the sequence of similar proteins isolated from different species. For example, one protein called "cytochrome c" has been examined in more than eighty species. These cytochrome c amino acid sequences represent "digital" bits of data that can be used to quantify differences between species, and these differences can be used to construct evolutionary trees much like those based upon comparisons of "analog" features of body structure. Such protein sequence trees—as well as trees based upon gene structure similarities—agree remarkably well with the evolutionary trees derived earlier from anatomic similarities. The agreement of evolutionary trees constructed from such completely different sorts of data has been taken by evolutionists as evidence of the validity of the intellectual framework on which the trees are based: the theory of evolution (see Jukes 1983, 1986).
The Creationist View on Species Similarities Leads to a Deadlock
However, creationists have an alternative interpretation of the amino acid sequence similarities reflected in the evolutionists' trees. They say that such sequence similarities in "related" species simply reflect the creator's choice to design similar species to function similarly, not only at the level of bones, muscles, and organs but also at the level of protein function—hence the amino acid sequence similarities.
Thus, the similarities between species in anatomy and protein structure can be interpreted in two entirely different ways. The evolutionists say that the similarity between features of, for example, humans and apes reflects the fact that these features were "copied" from a common ancestor; the creationists say that the two species were created independently but were designed with similar features so that they would function similarly. Both views seem consistent with the similarity data, but which view is correct?
A Possible Way to Resolve the Deadlock
One way to distinguish between copying and independent creation is suggested by analogy to the following true cases from the legal literature. In 1941, the author of a chemistry textbook was the plaintiff in a suit charging that portions of his textbook had been plagiarized by the author of a competing textbook. In 1946, the publisher of a trade directory for the construction industry made similar charges against a competing directory publisher. In both cases, mere similarity between the contents of the alleged copies and the originals was not considered compelling evidence of copying. After all, both chemistry textbooks were describing the same body of chemical knowledge and both directories listed members of the same industry, so substantial similarity would be expected even if no copying had occurred. However, in both cases errors present in the "originals" appeared in the alleged copies. The courts judged that it was inconceivable that the same errors could have been made independently by each plaintiff and defendant and ruled in both cases that copying had occurred. The principle that duplicated errors imply copying is well established in copyright law. (In recognition of this fact, directory publishers now routinely include false entries in their directories to trap potential plagiarizers.)
Can "errors" in modern species be used as evidence of "copying" from ancient ancestors? In fact, the answer to this question appears to be "yes," since recent molecular genetics investigations have uncovered some examples of the same "errors" present in the genetic material of humans and apes. To understand these findings it is necessary to know a little about deoxyribonucleic acid (DNA), the chemical molecule in which genetic information is stored.
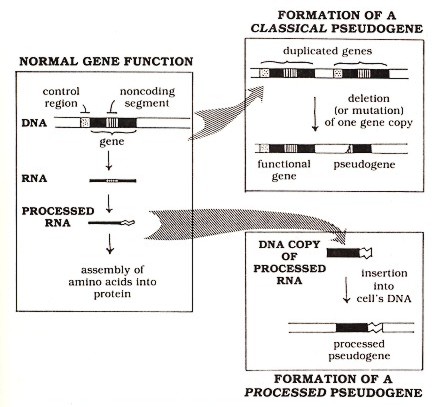 Figure 1: How genes function normally and how they give rise to pseudogenes
Figure 1: How genes function normally and how they give rise to pseudogenes
In one respect, the basic structure of DNA resembles that of proteins: both are made of linear chains of subunits. (Apart from this common feature, DNA and protein have many differences, which need not concern us here.) The subunits in DNA are called nucleotides, and the sequence of these nucleotides contains the genetic information. This information includes not only the "blueprint" specifying the sequence of amino acids in proteins but also various sorts of "punctuation"—control signals that ensure that the proteins are made in the proper amounts in the proper cells. The DNA acts somewhat indirectly in specifying protein structure. As diagrammed in the left panel of Figure 1 (above), this information is first copied into a molecule called ribonucleic acid (RNA). This initial copy of RNA undergoes several structural alterations, known collectively as processing. These alterations include the removal of unnecessary noncoding sequences from the RNA (the cross-hatched region in the figure) and some additions (represented by the wavy "tail" in the figure) that promote proper functioning of the RNA in the cell. It is the "processed" RNA that participates directly in the assembly of amino acids into proteins. The expression of a gene as an RNA copy is very tightly controlled, generally by highly specific regulatory sequences (represented in the figure by the stippled region) that occur in the DNA near the position where the RNA copy should begin but outside the copied region.
Recombinant DNA technology has in recent years allowed scientists to determine the sequence of nucleotides in segments of DNA from many species, and several million nucleotides' worth of information has accumulated. These sequences have vastly increased our understanding of how genes normally function; but, more to the point of this article, they have provided a treasure trove of genetic "errors" that are potential clues to the analysis of copying discussed earlier. In considering these "errors," I will focus upon two types of pseudogenes—that is, DNA sequences which are clearly related to known functional genes but which are apparently nonfunctional because of specific sequence alterations.
Pseudogenes: Genetic Errors of Two Kinds
The first type of pseudogene to be discovered (which I call the classical pseudogene) apparently arises from mishaps in a pattern of gene alteration that has been important to the development of normal functional genes: the pattern of duplication and differentiation. This pattern is evident from the frequent observation (in DNA from a variety of species) of blocks of sequences that have apparently been duplicated so that two or more repeats of similar sequences appear side by side. Presumably at the time of duplication each copy had an identical sequence. As DNA sequences are copied from generation to generation, mutations (mistakes in the normally accurate copying of DNA) can accumulate independently in the duplicated sequence copies. Some mutations may have no effect on the function of the gene. Others may lead to a protein that has a different function from that of the original gene. (Such differentiation of duplicated genes to develop new functions apparently accounts for a significant part of the expansion in complexity of the genes of higher organisms.) Finally, still other mutations, especially large deletions in the gene or alterations in the "punctuation" signals mentioned earlier, may completely destroy the function of a gene sequence and render it a pseudogene (see Figure 1, upper right panel).
The crucial defects in a pseudogene can often be recognized by comparing its sequence with that of the related functional gene. The kinds of mutations that destroy gene function are well known from studies of mutations that have disabled crucial nonduplicated genes, thereby causing genetic diseases. Such defective nonduplicated genes tend to disappear from populations over time because individuals lacking a functional copy of the gene are less capable of surviving to produce offspring. However, when a defective gene exists alongside a normal functioning copy, the abnormal sequence is usually harmless and may be perpetuated in the population as a pseudogene. Numerous pseudogenes of this type have been found in DNA from a variety of organisms, including humans.
An entirely different class of pseudogenes, known as processed pseudogenes, arises from naturally occurring insertions of extra gene copies into the cell's DNA. These inserted copies apparently derive from RNA molecules since they bear various features characteristic of the normal "processing" of RNA molecules-hence, the name processed pseudogenes. (see Figure 1, lower right panel. Note that the processed pseudogene lacks the noncoding region [cross-hatched segment] present in the original DNA and includes the "tail" sequence [wavy line] that was added to the RNA during processing. Both of these features indicate the derivation of the pseudogene from processed RNA.) Unlike classical pseudogenes, which are usually found close to the functional genes from which they are derived by duplication, processed pseudogenes are apparently inserted into DNA at random locations. This randomness is what one would expect for a sequence derived from an RNA molecule that can float freely away from its source gene (from which it was originally transcribed) before a copy is reinserted back into the DNA. Even if it encodes a correct amino acid sequence, a processed pseudogene is usually nonfunctional because it lacks the control sequences necessary for gene expression.
How Ancient Errors Can Persist in Modern Species
Each pseudogene that we observe is the result of a genetic accident that occurred in a single individual living at a particular time. A pseudogene arising in a muscle or liver cell of an individual would never leave those organs and would "die" when the individual died. In order for a pseudogene to be represented in later generations, one must assume that it arose either in one of the sex cells of the individual (egg or sperm) or early enough in embryonic development that it was present in the sex cells as they developed.
How could such a nonfunctional sequence, arising in a single individual, come to be preserved in all individuals of the species? A likely mechanism is that the pseudogene happened to lie close to an advantageous gene that became prevalent in a population by natural selection (the pseudogene "rode on the coat-tails" of the nearby advantageous gene). This mechanism for the establishment of a gene variant is most effective in very small populations in which a single dominant couple may supply most of the genes for the next generation. It is likely that pseudogenes arise with high frequency but we observe only those few that are preserved by unusual circumstances.
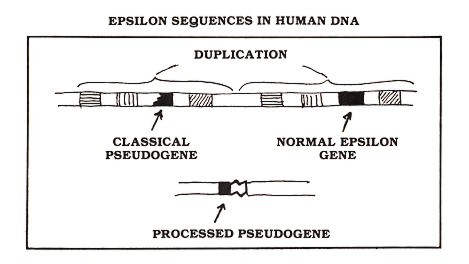 Figure 2: The human gene encoding the kind of antibody protein known as epsilon (black rectangle) gave rise to two pseudogenes—one classical and one processed. Both of these useless sequences are present in essentially every cell of your body
Figure 2: The human gene encoding the kind of antibody protein known as epsilon (black rectangle) gave rise to two pseudogenes—one classical and one processed. Both of these useless sequences are present in essentially every cell of your body
The extra burden of carrying along even a large pseudogene sequence—for example, 100,000 nucleotides—is insignificant for a mammalian cell with approximately three billion nucleotides' worth of information. There is, in any case, no known "proofreading" mechanism by which the cell might recognize and eliminate nonfunctional DNA. Functionless DNA sequences that experimenters have recently been able to insert into an organism's DNA are faithfully passed to descendants, and pseudogenes apparently behave similarly. The accumulation of functionless DNA is not completely uncompensated; deletions of DNA do occur, apparently as rare accidents that do not discriminate between functional and nonfunctional DNA. Deletions that remove crucial functional genes have been recognized as the cause of several genetic diseases, but other deletions that are harmless could remove some nonfunctional DNA. However, this is clearly an inefficient "garbage removal" mechanism, and, as an inevitable consequence of this inefficiency, substantial amounts of functionless "garbage" sequences have accumulated between the functional genes of most species. This is a surprising characteristic of the genetic material that was not appreciated until the past few years when recombinant DNA technology enabled molecular biologists to look beyond amino acid sequences to the structure of DNA itself.
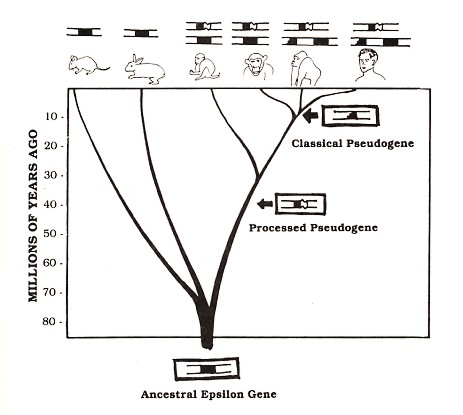 Figure 3: Our epsilon pseudogenes are shared by other species. The figure illustrates a few of the branches on our "family tree" for which the question of epsilon genes has been investigated. Each species is drawn below a diagrammatic representation of its epsilon-related sequences. According to the evolutionary point of view, branching points that are lower on this tree represent more ancient species divergence. In this diagram, humans are represented as being related more closely to gorillas than to chimpanzees; this view is controversial but consistent with the epsilon pseudogene data
Figure 3: Our epsilon pseudogenes are shared by other species. The figure illustrates a few of the branches on our "family tree" for which the question of epsilon genes has been investigated. Each species is drawn below a diagrammatic representation of its epsilon-related sequences. According to the evolutionary point of view, branching points that are lower on this tree represent more ancient species divergence. In this diagram, humans are represented as being related more closely to gorillas than to chimpanzees; this view is controversial but consistent with the epsilon pseudogene data
The Argument from DNA to Evolution: Shared Pseudogenes
The crucial observation relating the discovery of pseudogenes to the theory of evolution is this: some pseudogenes are shared between different species. As examples, let's focus upon two human pseudogenes which I studied with colleagues in Dr. Philip Leder's laboratory at the National Institutes of Health (Max et al., 1982; Battey et al., 1982). Similar results were obtained by Dr. Tasuku Honjo and colleagues in Japan, who extended their observations to a variety of primate species (Ueda et al., 1982; Hisajima et al., 1983; Ueda et al., 1985). My colleagues and I were studying the human gene encoding immunoglobulin epsilon (a kind of antibody protein that participates in allergic reactions). We found that, in addition to the expected functional gene, human DNA contains two epsilon pseudogenes—one processed and one classical (see Figure 2).
Evidence from our laboratory suggested that the processed epsilon pseudogene was inserted at the same spot in both human and chimpanzee DNA. Dr. Honjo's group investigated the DNA of other species and found evidence for this processed pseudogene in gorillas as well as several monkey species (see Figure 3). The classical pseudogene is found within a large block of duplicated genes (Flanagan and Rabbitts, 1982); the other genes in this block (hatched rectangles in Figure 2) are known to be functional, but one of the epsilon gene duplicates (black segments in Figure 2) suffered a deletion that removed DNA encoding about half of the amino acids of the epsilon protein, thereby completely disabling the gene. This pseudogene is apparently shared by man and gorilla but is not found in other apes or monkeys (see Figure 3). Other examples of shared pseudogenes are known (see for example, Chang and Slightom, 1984; Harris et al., 1984), and additional examples will almost certainly come to light as human and other mammalian DNAs are studied. But even a single example is sufficient to make a strong argument against the creationist viewpoint.
This argument can be understood by analogy with the legal cases discussed earlier in which shared errors were recognized as proof of copying. The appearance of the same "error"—that is, the same useless pseudogene in the same position in human and ape DNA—cannot logically be explained by independent origins of these two sequences. The creationist argument discussed earlier—that similarities in DNA sequence simply reflect the creator's plans for similar protein function in similar species—does not apply to pseudogenes because these sequences do not encode any functional protein. The possibility of identical rare genetic accidents creating the same two pseudogenes in ape and human DNA by chance is so unlikely that it can be dismissed. As in the copyright cases discussed earlier, these shared "errors" indicate that copying of some sort must have occurred. Since there is no mechanism by which sequences from modern apes could be copied into human DNA, or vice versa, the existence of the two shared pseudogenes leads to the logical conclusion that both the human and ape sequences were copied from ancestral pseudogenes that must have arisen in a common ancestor of humans and apes.
Extensions of the Shared Pseudogene Argument
This evidence for a common ancestor clinches the argument for evolution that follows from the shared epsilon pseudogenes. These pseudogenes link only humans and apes on the evolutionary family tree, but it is obvious that other shared pseudogene data can potentially be used to support other branches of the tree. By similar logic, other functionless features of DNA which are shared by two species and which are too complex or too specific to have occurred independently by chance may be taken as evidence of common ancestry. Examples of such features already known to be shared between humans and chimpanzees include several other types of pseudogenes and the occurrence of other inserted sequence elements at the same location in the DNA of both species.
Another entire article could be written on these "other inserted sequence elements." Such elements occur with impressive variety in many species (see, for example, Kuff et al., 1983; Rogers, 1985; Weiner et al., 1986) and include sequences resembling retroviruses, which are known to insert their DNA into the DNA of cells that they infect. Two well-known examples of retroviruses are the pathogenic viruses causing AIDS and feline leukemia, but our DNA contains "endogenous" retroviral sequences that are apparently harmless. One such endogenous retroviral sequence, apparently "caught" by an ancestor of ours millions of years ago, is now found embedded at the same position in human and chimpanzee DNA (Bonner et al., 1982).
Evolutionists as early as Darwin pointed to vestigial structures—such as the functionless eyes of blind cave-dwelling animals or the rudimentary pelvic bones of some snakes—as supporting the evolutionary viewpoint. These structures serve no apparent purpose that could explain their design by a creator but can easily be understood in the evolutionary perspective as deriving from functional structures in ancestral species. Vestigial genetic sequences—that is, pseudogenes—provide exquisite examples of vestigial structures and, thus, especially compelling evidence for evolution. In contrast to some proposed vestigial organs, they can be studied in a variety of species, their relationship to their functional counterpart is obvious, and, especially for the processed pseudogenes, they can be assumed to be totally functionless from the instant of their creation (some organs cited as vestigial, for example, the human appendix, have been argued to have some function).
Absolute Proof? Science Can Advance Without It.
Do the shared pseudogenes prove that humans and apes had a common ancestor? Actually, no scientific knowledge is based upon unassailable proof of the sort that supports mathematical theorems. Instead, science advances by the accumulation of clues sought by persistent detectives (scientists) who try to derive logical and unbiased deductions from these clues. Like a jury presented with these clues, we can try to arrive at the most likely verdict even though we recognize that our facts are incomplete; there are no living "witnesses" to the eons of evolution, so we must do the best we can from the clues at hand. In the "case of shared pseudogenes," an unbiased jury would surely conclude that copying from a shared ancestor was the most likely explanation, consistent with the evolutionary interpretation. This conclusion would follow the logic of the actual legal principal of copyright litigation regarding shared errors as evidence of plagiarism, as discussed earlier.
One feature of science that distinguishes it from revealed religious belief (and evolutionists from creationists) is the scientific conviction that new knowledge about the past can be obtained from thoughtfully designed analysis of the real world. Creationists often claim that, since the origin of species occurred in the distant past, there is no scientifically valid way to study the process today and so evolution is not real science testable by experiment. However, even without actual experiments, a scientific hypothesis can be tested if it suggests a nontrivial prediction that can be verified, or falsified, by the collection of more data. Indeed, the interpretation of shared pseudogenes outlined here represents a hypothesis that can be tested because it suggests a rather startling implication: from a comparison between two nucleotide sequences from a single species—that is, the sequences of a processed pseudogene and of the functional gene from which it derived—it should be possible to predict accurately which other species will share the same pseudogene and which will not. To understand the logic of such a prediction, consider the fact that a newly formed processed pseudogene exists only in the species in which it arose, while an "old" processed pseudogene that arose in an ancient species should be found in modern descendants of that species. Thus, according to the evolutionary model, if we knew when a processed pseudogene arose and could thus fix its origin to a particular position on , the accepted evolutionary "tree," we would predict that the same processed pseudogene should be found in modern species that derive from that point on the tree and not in any other branches. In fact, there is a way to estimate when a given processed pseudogene was formed. It turns out that "silent" mutations—that is, mutations that have no effect on the survival of the organism (like mutations in useless pseudogenes)—accumulate at a fairly uniform rate. This rate has been estimated by examining the number of "silent" sequence differences between corresponding functional genes in two species and by comparing this number with the approximate date of divergence of the same two species as indicated by the fossil record. Given this mutation rate and the number of sequence differences between a processed pseudogene and its fun, ctional source gene from the same species, one can estimate the date of origin of the pseudogene; then, using this date, one can derive predictions about appearance of the pseudogene in other species on the evolutionary family tree.
Consider, for example, the processed human epsilon pseudogene discussed earlier. The number of differences between this pseudogene and the corresponding sequence of the human functional epsilon gene suggests that this pseudogene arose about 40 million years ago. Therefore, the interpretation of processed pseudogenes described above would predict that mice and rabbits (which are thought to have diverged from the human lineage 70 to 80 million years ago, before the apparent origin of the pseudogene) should not carry the pseudogene, while apes and Old World monkeys (whose estimated dates of divergence from the human lineage [5 to 10 million and 30 million years ago, respectively] are both after the apparent pseudogene origin) should carry the pseudogene in their DNA. Available evidence confirms all of these predictions (see Figure 3) and is also consistent with the evolutionary interpretation for the case of several other known processed pseudogenes (see, for example, Anagnou et al., 1984). More shared processed pseudogenes will certainly be discovered, and only time will tell how consistently such predictions are confirmed. Repeated instances of this kind of prediction and confirmation can supply convincing evidence for evolution even though some kinds of direct experiments to test evolution, such as experiments involving living dinosaurs, are impossible.
Conclusion
As new examples of shared pseudogenes are discovered by molecular geneticists, this information will surely join the immense body of clues from other disciplines which, collectively, already provide overwhelming evidence for evolution. Despite this impressive evidence, no scientist believes that all the answers are in on evolution or that our current understanding of pseudogenes is immune from revision in light of future knowledge. Indeed, scientists in laboratories throughout the world are continuing to probe the genes of various species, comparing the molecular genetics data with the fossil record and refining our knowledge of our species' history.
At the present stage of this never-ending research, the evidence suggests what to me is an awesome notion: like a biological Rosetta Stone or Dead Sea Scroll, our own DNA—an Encyclopaedia Brittanica's worth of information in every cell of the body—contains a record of the past which we are just now learning to read. This record, reflecting millions of years of genetic history, includes the relics of ancient genetic accidents that occurred before our apelike ancestors roamed the plains of Africa, relics that we now share with other descendants of the same ancestors—the great apes.
Acknowledgements
I am grateful to the many colleagues and friends who read earlier versions of this manuscript and made suggestions for improving clarity and balance. The comments of John Immerwahr, Jonathan Silver, and Mary Duncan were especially helpful.
