Explore Evolution completely ignores studies showing that mutations in both protein coding sequences and in non-coding cis-regulatory element sequences (CREs) are responsible for changes in morphology. Explore Evolution muddies the distinction between mutations which affect protein structure and function, and mutations which affect when and where genes are turned on or off.
In its discussion of DNA and mutations, Explore Evolution asserts:
DNA is actually closer to the bottom of the organizational ladder. Yes, it directs the building of proteins, and yes, proteins are important. But DNA does not direct how the overall body plan gets built.p. 110
In making this claim, Explore Evolution fails to acknowledge the extensive research on mutations in DNA sequences that do not encode proteins, but which have important morphological effects. Explore Evolution thereby misses out on the opportunity to present the genuinely relevant and interesting scientific debate in evolutionary morphology concerning the relative importance of mutations in protein-coding sequences versus mutations in non-coding sequences.
An important tenet of evolutionary developmental biology ("evo devo") is that adaptive mutations affecting morphology are more likely to occur in the cis-regulatory regions than in the protein-coding regions of genes Although this claim may be true, it is at best premature. Adaptation and speciation probably proceed through a combination of cis-regulatory and structural mutations, with a substantial contribution of the latter.Hoekstra et al., (2007) "The locus of evolution: Evo devo and the genetics of adaptation," Evolution 61:005
Non-coding regions of DNA contain genetic switches called cis-regulatory elements (CREs). CREs control when and where a gene is turned on (expressed). Mutations in the CREs of Ultrabithorax are responsible for that gene being turned off in fly hindwing resulting in the four-winged fruit fly. As Sean Carroll and colleagues argue, recent analyses have shown that mutations in CREs play an important role in morphological evolution.
A growing number of case studies exploring the mechanisms of morphological changes have provided direct evidence that CRE evolution plays a major role. From these examples, we have identified general rules regarding regulatory evolution, namely how regulatory evolution exploits available genetic components, irrespective of their hierarchical position in gene networks to generate novelty, and minimizes fitness penalties. These rules offer a rationale explaining why regulatory changes are more commonly favored over other kinds of genetic changes in the process of morphological evolution, from the simplest traits diverging within or among species to body-plan differences at higher taxonomic levels.Prud'homme et al., (2007), "Emerging Principles of Regulatory Evolution", PNAS 104:8611
An example of the Pitx1 gene in the marine stickleback fish is shown below. Genetic studies of sticklebacks by David Kingsley and colleagues have established the mutations in Pitx1 have played an important role in the evolution of sticklebacks. There two important CREs controlling Pitx1 expression. One CRE is postulated to control Pitx1 expression in the mouth and jaw while another CRE is postulated to control Pitx1 expression in the pelvic hindlimb.
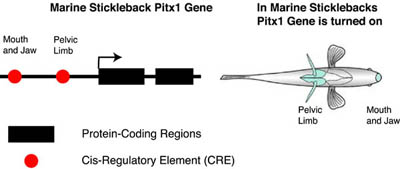
Mutations in the protein-coding region can affect the sequence and the function of a protein. On the other hand, mutations in the CRE can affect when and where a gene is turned on. Have mutations in CREs or in protein-coding regions of genes been involved in morphological evolution? This question has been addressed in a number of different studies described below.
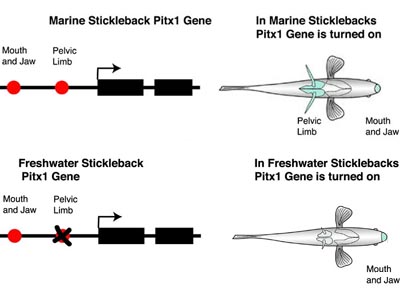
David Kingsley and colleagues (Shapiro et al., 2003) have studied the evolution of sticklebacks. Over the last 10,000 years, marine sticklebacks have invaded freshwater lakes and have undergone an extensive adaptive radiation. One important adaptation in freshwater stickleback has been the reduction of pelvic spines that helps to reduce predation from arthropods. To address what gene(s) are responsible for pelvic spine reduction, Kingsley and colleagues conducted genetic experiments on sticklebacks and showed that mutation in the Pitx1 gene was responsible.
Recall that Pitx1 is expressed in development of the pelvic spine and of the mouth and jaws in marine sticklebacks. However, in freshwater sticklebacks with reduced pelvic spines, Pitx1 is expressed only in the mouth and jaws. So, although they have not yet identified the precise mutation(s), a technically demanding job of finding a needle in a haystack, this observation strongly suggests that mutations in the CRE controlling pelvic spine expression of Pitx1 are responsible for this reduction. Note that a mutation in the protein-coding region of Pitx1 would affect Pitx1 function in both the mouth, jaw and pelvic spines. This type of mutation is fatal in mice. However, because mutations in one CRE do not affect the function of a different CRE, Pitx1 function can be lost in the pelvic spine and retained in the mouth and jaw of freshwater sticklebacks.
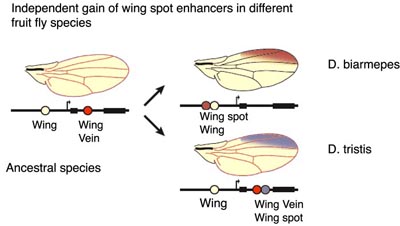
As Kingsley and colleagues show, the mutations in a CRE can cause the loss of Pitx1 function in pelvic spines, but can mutations generate new CREs? Sean Carroll and colleagues (Prud'homme et al., 2006) have addressed this question by focusing upon how wing spots have evolved in fruit flies. Wing spots play an important role in mating behaviors of fruit flies and would be predicted to be under strong natural selection. By a functional analysis of wingspot CREs of different fruit fly species, Prud'homme et al. show that CREs responsible for wing spots in fruit flies have been independently gained as shown below. Interestingly, these new CREs have been generated by the modification of pre-existing CREs.
David Stern and colleagues have examined a different type of morphological evolution in fruit flies, trichome formation on limbs. They have recently discovered that multiple mutations in the CREs controlling the genetic toolkit gene, shavenbaby, are responsible for the evolution of this morphological feature.
Here we examine the genetic basis of a trichome pattern difference between Drosophila species, previously shown to result from the evolution of a single gene, shavenbaby (svb), probably through cis-regulatory changes(6),,, Our results demonstrate that the accumulation of multiple small-effect changes at a single locus underlies the evolution of a morphological difference between species. These data support the view that alleles of large effect that distinguish species may sometimes reflect the accumulation of multiple mutations of small effect at select genes.McGregor et al., "Morphological evolution through multiple cis-regulatory mutations at a single gene," (2007), Nature 448:487
Mutations in CREs in fruit flies and sticklebacks have been shown to play important roles in their morphological evolution. Do mutations in CREs play any role in the evolution of humans? To address this question, Greg Wray and colleagues examined the PDYN gene which is involved in the synthesis of endogenous brain opiates. Through comparing the genomes of chimps and humans, population genetics and experimental analyses, they demonstrate that a 68 base-pair element is important for turning the PDYN gene on at higher levels in human brains and that this element has been under positive natural selection.
Changes in the cis-regulation of neural genes likely contributed to the evolution of our species' unique attributes, but evidence of a role for natural selection has been lacking. We found that positive natural selection altered the cis-regulation of human prodynorphin, the precursor molecule for a suite of endogenous opioids and neuropeptides with critical roles in regulating perception, behavior, and memory.Rockman et al., "Ancient and Recent Positive Selection Transformed Opioid cis-Regulation in Humans," (2005), PLOS Biology 3, p. 387
This is an important and active field of evolutionary research, and if Explore Evolution aimed to live up to its title, it is a field that could have generated the basis for genuine inquiry. Instead, unsolved questions are offered as unsolvable obstacles, and ongoing scientific research is ignored or obscured to serve the authors' misconceptions.
