Explore Evolution wrongly state that biologists originally maintained that the genetic code is absolutely universal (invariant); that this absolute universality was considered evidence for common descent; that this would be a reasonable inference because changing the code would be invariably lethal ("not survivable"); and finally, that the claim of universality fell apart in the 1980s with the discovery of variant genetic codes. Thus, the authors claim, the genetic code is not universal and the inference of common descent is in question and life must have "multiple separate origins." They cite physicist Hubert Yockey to justify the claim: "Some scientists think this is a possibility, saying that the evidence may point to a polyphyletic view of the history of life." (p. 59)
There are many problems with this argument, which is based on misunderstanding and misrepresentation of the available knowledge and of the scientific record.
First, contrary to the key assertion, scientists have been aware of natural genetic code mutants since at least the 1960s, and the actual molecular mechanism of some of these mutations (such as "suppressors of amber") was elucidated in both bacteria and yeast (Goodman HM, Abelson J, Landy A, Brenner S, Smith JD. (1968) "Amber suppression: a nucleotide change in the anticodon of a tyrosine transfer RNA." Nature 217:1019-24; Capecchi MR, Hughes SH, Wahl GM. (1975) "Yeast super-suppressors are altered tRNAs capable of translating a nonsense codon in vitro." Cell 6:269-77.) Amber suppressor mutations change the read-out of certain codons from STOP to an amino acid by altering the structure of one of the transfer RNAs. This tRNA recognizes the codons in messenger RNAs and allows the addition of the correct amino acid during protein synthesis. These mutants showed how new variant genetic codes can evolve, and what kind of selective pressures can favor such changes (in this case, the need for reversion of point mutations which introduce deleterious STOP codons in critical genes). Therefore, it was recognized fairly early that the genetic code did not need to be absolutely invariant to be fundamentally shared between all organisms ("universal"). Already in 1966, Francis Crick stated in his Croonian lecture:
The best evidence to date [for the universality of the code] is probably the excellent agreement between the code deduced for E. coli and the mutagenic data derived from tobacco plants or human beings. There is thus little doubt that the genetic code is similar in most organisms. Whether there are any organisms which use a slightly modified version of the code remains to be seen.Crick, FH (1967) The Croonian Lecture. "The Genetic Code." Proc R Soc Lond B Biol Sci. 167:331-47
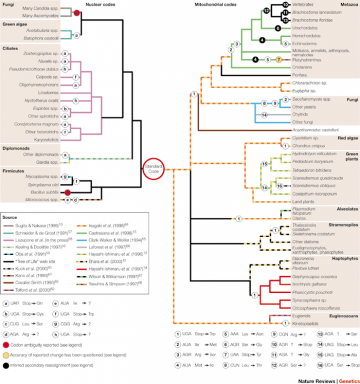 Code Variants: from Knight RD, Freeland SJ, Landweber LF. "Rewiring the keyboard: evolvability of the genetic code." Nat Rev Genet. 2001; 2:49-58
Code Variants: from Knight RD, Freeland SJ, Landweber LF. "Rewiring the keyboard: evolvability of the genetic code." Nat Rev Genet. 2001; 2:49-58 Second, the small number of organisms with variant genetic codes and the limited extent of the changes (involving a few codons at most) strongly support the view that these represent new variations of the "standard," universal code, as opposed to independently originated codes. Moreover, the known code variants themselves offer in many cases evidence for common descent, being shared by related organisms according to the established phylogenetic hierarchy, as shown in the figure below, from Knight RD, Freeland SJ, Landweber LF. (2001) "Rewiring the keyboard: evolvability of the genetic code." Nat Rev Genet. 2:49-58, which contains a thorough discussion on the phylogenetic distribution and mechanisms of genetic code variation.
In particular, the authors of Explore Evolution mention the specific example of organisms in which 2 of the 3 "stop codons" have been reassigned to encode for amino acids (change "a" in the figure above). They argue:It's very hard to see how an organism could have survived a transformation from the standard code to this one. Changing to this new code would cause the cell to produce useless strings of extra amino acids when it should have stopped protein production.Explore Evolution, p. 58
However, the mechanisms that underly this particular kind of code change in some organisms are known, and undermine the authors' argument. The studies have been performed mainly in Ciliates, a group of unicellular eukaryotes belonging to the Protozoans, which include Tetrahymena, the organism specifically cited by the authors. Notably, Ciliates have a peculiar genomic organization, with hundreds of very small chromosomes, often containing a single gene, organized in two distinct nuclei; moreover, their genes tend to have unusually short sequences past their termination codons. This is important because it means that mutations that suppress termination (such as that mentioned by the authors) are less likely to generate very long amino acid stretches past the normal protein end, and hence to cause deleterious phenotypes. Consistent with this possibility, Ciliates comprise the majority of organisms with alternative genetic codes containing termination suppressors (Knight RD, Freeland SJ, Landweber LF. (2001) "Rewiring the keyboard: evolvability of the genetic code." Nat Rev Genet. 2:49-58; Lopuzone CA, Knight RD, Landweber LF. (2001) "The molecular basis of genetic code change in ciliates." Curr Biol. 11:65-74).
The particular genetic code mentioned by the authors, in which the UAG and UAA codons are used to encode the amino acid glutamine instead of STOP, results from two sets of changes. The first involves a reassignment of the transfer RNA for glutamine to recognize UAG and UAA. Interestingly, this kind of change can occur through intermediates with only partial effects ("wobble") (Schultz DW, Yarus M. 1994 "Transfer RNA mutation and the malleability of the genetic code" J. Mol. Biol. 235(5):1377-80). The second set of changes affects eRF1, one of the proteins involved in recognizing STOP codons (Lopuzone CA, Knight RD, Landweber LF. 2001 "The molecular basis of genetic code change in ciliates." Curr Biol. 11:65-74). Because of this, it is not "very hard" at all, and in fact very possible, to envision gradual evolution of this new genetic code through intermediates in which the codon interpretation is ambiguous; "hybrid codes" in a sense.
To justify the claim that some scientists see variation in the genetic code as evidence against a single tree of life, the authors quote a sentence from a 1992 book by Hubert Yockey, a physicist with an interest in information theory of biological systems. Interestingly, the quote in question is absent from the latest edition of the book. More importantly, in the current edition, Yockey approvingly quotes Francis Crick's suggestion that all extant life forms descended from a small interbreeding population (i.e. common descent), and once again prophesizing the possibility of organisms with variant codes:
Crick (1981), in one of those marvelous intuitions that have led him to so many discoveries, and without the mathematical argument above, has proposed: What the code suggests is that life, at some stage, went through at least one bottleneck, a small interbreeding population from which all subsequent life has descended Nevertheless, one is mildly surprised that several versions of the code did not emerge, and the fact that the mitochondrial codes are slightly different from the rest supports this.Yockey H, (2005) Information Theory, Evolution, and The Origin of Life. Cambridge University Press, 2005, p.102
Here, the authors of Explore Evolution make the fundamental mistake of conflating (either on purpose or due to a misunderstanding of the underlying issues) two very different questions: common descent, whether extant organisms can trace their ancestry to a single population at some point in the past, and abiogenesis, whether life originated only once. Quite obviously the two issues are distinct. It is entirely possible that life originated more than once, but that early life forms were so promiscuous in sharing genetic material that they constituted, from a genetic standpoint, a single population from which all later organisms evolved. Essentially no scientist at this point objects to the possibility of the latter proposition, although some disagree as to the pattern in which various lineages arose from the original population. These differences may affect the extent to which linear, vertical descent lineages can be unequivocally identified when analyzing the deepest phylogenetic relationships (such as the separation of life into the Domains of Eubacteria, Archaea, and Eukarya), but do not change the view that all organisms ultimately are phylogenetically related.
