To buttress the false claim that developmental pathways regulated by Hox genes cannot evolve Explore Evolution engages in a major error of omission by failing to address the issue of modularity. This is concept is not obscure - a PubMed search of "modularity and evolution" yields over 140 publications in the last ten years. Modularity describes the independent control of hierarchical levels in development and evolution ranging from anatomical structures to signaling pathways to genetic switches.
In 1996, modularity in development and evolution was fully discussed in the influential book on evolutionary developmental biology, The Shape of Life by Rudolf Raff. As a book review of the Shape of Life in the journal Bioscience notes:
Raff defines modules as units " that are distinct in genetic specification, autonomous features, hierarchical organization, interactions with other modules, location, time of occurrence, and dynamic properties" Modularity permits change because developmental processes are free to dissociate, duplicate, diverge, and be co-opted to new uses. Although developmental modules themselves are complex, Raff argues that they may often be activated by single or few regulatory genes acting as switches, such as Pax-6 in eye development. The mostly likely changes in ontogeny are those that alter the relative timing, number, or location of preexisting modules rather than those that produce wholly novel modules or structures. This feature of development also helps to explain the essential conservation of body plan.Terri Williams, "The Modularity of Development" (1998), Bioscience 48 p. 60
As Eric Davidson notes in 2006, the modularity of cis-regulatory elements (CREs) is now well-established.
When cis-regulatory sequences were first proposed to be generally modular in organization (Kirchamer et al. 1996), there were on ly a modest number of examples from work in Drosophila and sea urchins, and the idea was largely inferential. Now there are literally scores of genes for which detailed experimental analyses have demonstrated sharply modular cis-regulatory elements, such that given non-overlapping regions of the genomic DNA each control a specific subcomponent of the overall expression pattern.Eric Davidson, The Regulatory Genome: gene regulatory networks in development and evolution (2006) p. 33
How does the modularity of CRE's relate to whether the integrated networks regulated by Hox genes can evolve? Consider how Ubx regulates the target genes Sal and CG13222 by binding to their CREs. A mutation that disabled Ubx binding to CG13222 would prevent it from being from turned on in the hindwing, but would not affect Ubx turning off Sal in the hindwing. Similarly, a mutation in Omb that caused it to turned off in the hindwing would not affect how Ubx controls Sal or CG13222.
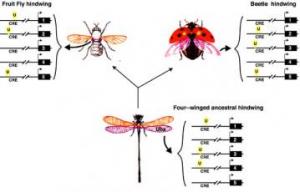 Hindwing Evolution
Hindwing Evolution Thus Ubx binding sites on CREs of different genes can be gained or lost independently of binding sites for other proteins genes. Although these genes can be involved in a complicated gene network, the modularity of CREs allow specific genes to be independently changed without affecting the rest of the network. Ubx is known to regulate hindwing development in all insects, from beetles to fruitflies, even though the morphology of their greatly differs. These major changes in morphology are easily explained when considering how Ubx binding sites on CREs can evolve.
